Objectives ⬇
- Clarify the differences between evidence-based practice and translational research.
- Explore the potential contributions of bioinformatics and computational biology to evidence for practice.
- Explore models for introducing research findings into practice.
- Assess barriers to research utilization in practice.
Key Terms ⬆ ⬇
Introduction ⬆ ⬇
Mr. James is an 87-year-old man with osteoarthritis in his knees. He is frail and very thin and requires assistance getting out of bed. Mary, a new registered nurse, is making her rounds with her team members and nurse's aide. Realizing Mr. James is at risk for skin breakdown and falls, she reviews the agency policy manual regarding pressure ulcer and fall prevention. Which other resources could Mary consult if she wanted more information on preventing these issues? If Mary wanted to know what the current research suggests about preventing each of these conditions, how would she obtain this information?
This chapter introduces the concept of translational research and its role in evidence-based practice (EBP) with a specific emphasis on nursing informatics and bioinformatics. Before pursuing the content in this chapter, the reader should already understand nursing research, the Foundation of Knowledge model, and knowledge generation through nursing research. Next, key words and definitions used in this chapter are briefly described. Classic sources (5 years or older) are used to enhance the reference base and give the reader a sense of the evolution of translational science and EBP.
Clarification of Terms ⬆ ⬇
Evidence-based practice, translational research, and research utilization are terms that have been used to describe the application of evidential knowledge to clinical practice. The following paragraphs explore the definitions of each term. Although these terms are related, they have slightly different meanings and applications.
Evidence-based practice (EBP), developed originally for its application to medicine, was classically defined by Sackett et al. (1996) as “[t]he conscientious, explicit and judicious use of current best evidence in making decisions about the care of individual patients” (p. 71). The “best evidence” in this context refers to more than just research. Goode and Piedalue (1999) stated that EBP should be combined with other knowledge sources and “involves the synthesis of knowledge from research, retrospective or concurrent chart review, quality improvement and risk data, international, national, and local standards, infection control data, pathophysiology, cost effectiveness analysis, benchmarking data, patient preferences, and clinical expertise” (p. 15). EBP starts with a clinical question to resolve a clinical problem (Figure 23-1). For example, published research studies are used in healthcare quality initiatives as the evidence behind the development of practice algorithms designed to decrease practice variability, increase patient safety, improve patient outcomes, and eliminate unnecessary costs. Use of EBP promotes the use of clinical judgment and knowledge in relation to the patient's contextual situation and preferences, with procedures and protocols being linked to scientific evidence rather than based on what is customary practice or opinion. Weiss et al. (2018) suggested that “the focus of EBP is the systematic process of review, critique, and synthesis of research evidence and relevant sources of nonresearch evidence to develop a best practice protocol incorporating logistical considerations for implementation in the local context” (p. 427). The relevant sources of nonresearch refer to organizational values, local data, local context, and patient preferences.
Figure 23-1 Evidence-Based Practice
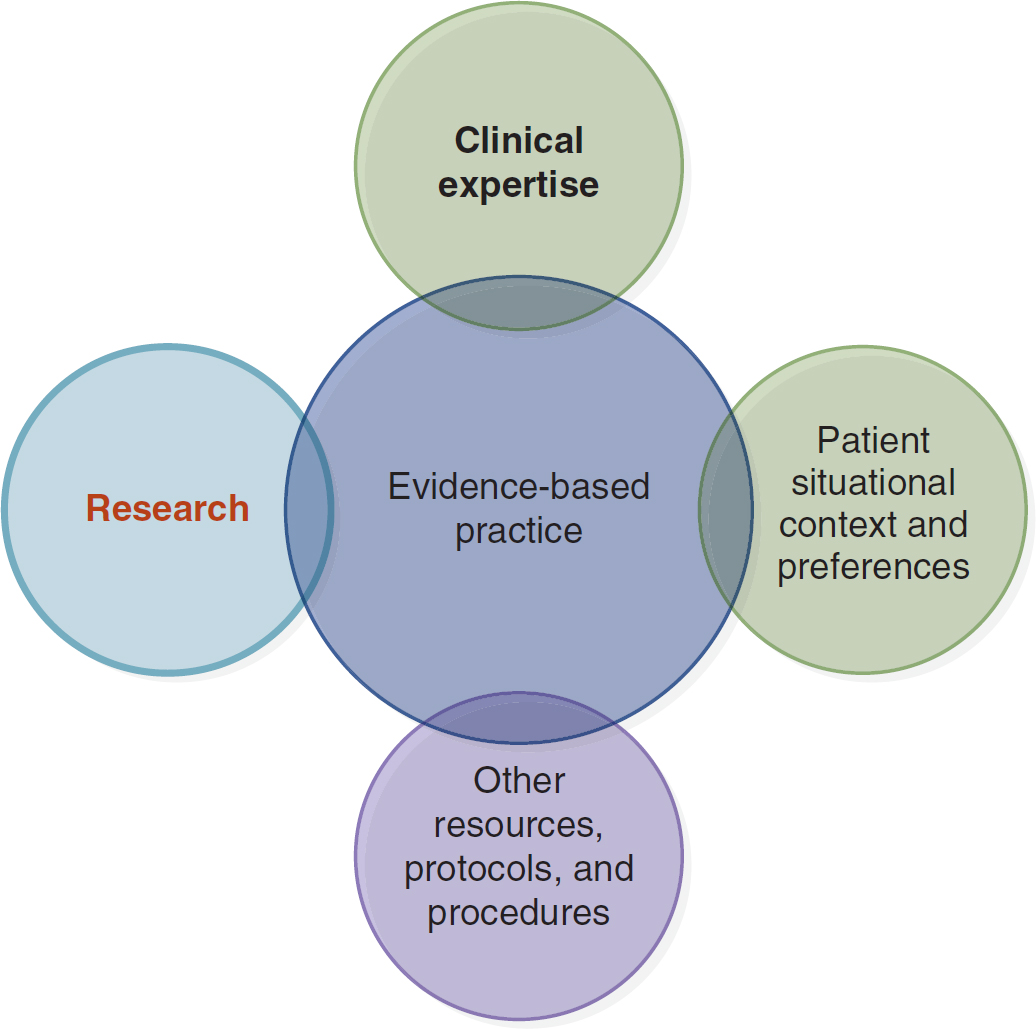
Evidence-based practice comprises research, clinical expertise, patient situational context and preferences, and other resources, protocols, and procedures.
Research utilization is the use of findings from one or more research studies in a practical application unrelated to the original study (Polit & Beck, 2008, p. 29), resulting in the generation of new knowledge. Stetler (2001) defined research utilization as the “process of transforming research knowledge into practice” (p. 274). Research utilization can be self-limiting if research is inconsistent or not enough research is available to develop a consensus regarding the answer to the clinical question (Kirchhoff, 2004). Squires et al. (2011) discussed four distinct types of research utilization: overall use; conceptual use, where research prompted a change in thinking but not necessarily practice; symbolic (persuasive) use, where research was used to support a position or influence the position of another; and the direct application of research findings in practice (instrumental use).
Translational research refers to the methods used in translating medical, biomedical, informatics, and nursing research into bedside clinical interventions. As depicted in Figure 23-2, new things, new ideas, new techniques, and new approaches must progress through several stages before they are widely implemented in practice. Woolf (2008) described translational research in two ways:
Figure 23-2 Translational Research Pathways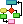
A flowchart depicts the progression of creation, assessment, uptake, and spread through the introduction of new elements like things, ideas, techniques, and approaches, which result in invention, evaluation, adoption, and diffusion respectively.
- T1: The transfer of clinical research to its first testing on humans
- T2: The transfer of clinical research to an everyday clinical practice setting
The National Center for Advancing Translational Sciences (NCATS, 2021) developed a translational science spectrum that expands on Woolf's original description of progress toward clinical application. The NCATS spectrum includes basic research, preclinical research, clinical research, clinical implementation, and public health. “At all stages of the spectrum, NCATS develops new approaches, demonstrates their usefulness and disseminates the findings. Patient involvement is a critical feature of all stages in translation” (para. 1).
Difficulties in translating research to the clinical practice setting exist when research applications do not fit well within the clinical context or when practical considerations constrain the application in a clinical setting. Translational research is complicated by the follow-up analysis, practice, and policy changes that occur when adopting research into practice; consequently, available healthcare EBPs often are not fully incorporated into daily care (Titler, 2004, 2010). As Aarden et al. (2021) pointed out in their examination of the translational lag narrative, “basic science rapidly produces new insights into the biological determinants of human health and disease, but clinical innovation fails to deliver new tools to improve patient care at a comparable pace” (p. 2). Organizational culture influences the changes made to a clinical application and establishes the groundwork and the support for change-making activities (Titler, 2004). The study of ways to promote the adoption of evidence in a healthcare context is called translation science (Titler, 2010). NCATS (2019) of the National Institutes of Health (NIH) provided this definition of translation: “[t]he process of turning observations in the laboratory, clinic and community into interventions that improve the health of individuals and the public-from diagnostics and therapeutics to medical procedures and behavioral changes” (para. 2). Access this center at https://ncats.nih.gov.
Translational bioinformatics is the “development of storage, analytic, and interpretive methods to optimize the transformation of increasingly voluminous biomedical data and genomic data, into proactive, predictive, preventive, and participatory health” (American Medical Informatics Association [AMIA], n.d.-c, para. 1). It integrates biological and clinical data and the evolution of clinical informatics methodology to include biological observations. “The end product of translational bioinformatics is newly found knowledge from these integrative efforts that can be disseminated to a variety of stakeholders, including biomedical scientists, clinicians, and patients” (AMIA, n.d.-c, para. 1).
Clinical informatics is the “application of informatics and information technology to deliver healthcare services. It is also referred to as applied clinical informatics and operational informatics” (AMIA, n.d.-a, para. 1).
Clinical research informatics is defined by AMIA (n.d.-b) as involving the use of informatics in the discovery and management of new knowledge relating to health and disease. “It includes management of information related to clinical trials and also involves informatics related to secondary research use of clinical data. Clinical research informatics and translational bioinformatics are the primary domains related to informatics activities to support translational research” (para. 1).
History of Evidence-Based Practice ⬆ ⬇
Research results are crucial to furthering EBP. The concept of using randomized controlled trials (RCTs) and systematic reviews as the gold standard against which one should evaluate the validity and effectiveness of a clinical intervention was introduced in 1972 by Archie Cochrane (1972), a scientist and physician. Cochrane's experiences as a prisoner of war and medical officer while interning during World War II led to his belief that not all medical interventions were needed and that some caused more harm than good. Cochrane viewed the RCT as a means of validating clinical interventions and limiting the interventions to those that were scientifically based, effective, and necessary (Dickersin & Manheimer, 1998). As explanatory information, an RCT involves testing an intervention in a study with two randomly selected groups, one receiving the intervention and the other serving as a control group. A systematic review is a collection and analysis of research on a particular topic (more on this later).
Cochrane's colleague, Iain Chalmers, began compiling a comprehensive clinical trials registry of 3,500 clinical trial results in the field of perinatal medicine. In 1988, after being published in print 3 years earlier, the registry became available electronically. Chalmers's methods for compiling the trials database became a model for future registry assembly. Eventually, the National Health Service in the United Kingdom, recognizing the value of and need for systemic reviews for all of health care, developed the Cochrane Center. The Cochrane Collaboration was initiated in 1993 and expanded internationally to maintain systematic reviews in all areas of health care (Dickersin & Manheimer, 1998). Many universities subscribe to the Cochrane Collaboration database, making this information easily accessible to students, faculty, and nurses who work for university hospital systems. Visit www.cochrane.org for the latest Cochrane evidence. Cochrane announced in 2019 its intention to establish a new Cochrane network across the United States. Access the U.S. network here: http://US.cochrane.org
Evidence ⬆ ⬇
The RCT is considered the most reliable source of evidence. Yet RCTs are not always available; consequently, nurses must use critical analysis using the best available evidence on which to base their clinical decision-making (Baumann, 2010). The updated Stetler (2001) model of research utilization identified internal and external forms of evidence. External evidence originates from research and national experts, whereas internal forms of evidence originate from nontraditional sources, such as clinical experience and quality improvement data.
Evidence includes standards of practice, codes of ethics, philosophies of nursing, autobiographical stories, esthetic criticism, works of art, qualitative studies, and patient and clinical knowledge (Melnyk et al., 2000). French (2002) summarized evidence as “truth, knowledge (including tacit, expert opinion and experiential), primary research findings, meta-analyses and systematic reviews” (p. 254). Brown (2013) provided this definition of evidence: “Objective knowledge or information used as the basis for a clinical protocol, clinical decision, or clinical action. Evidence sources include research, agency data regarding system performance and patient outcomes, large healthcare databases, and expert opinion” (p. 439). Nurses may additionally draw on evidence from the context of care, such as audit and performance data, the culture of the organization, social and professional networks, discussion with stakeholders, and local or national policy (Rycroft-Malone et al., 2004, p. 86).
Concern has been voiced by nurse theorists that nurses are being influenced too much by the medical model in accepting the RCT as the only true source of evidence, thereby “reverting to the medical perspective” rather than incorporating “theory-guided evidence and diverse ways of knowing” (Fawcett et al., 2001, p. 115). The context change from medicine to nursing requires nurses to apply other knowledge and nursing theory. The use of research results as the sole basis for clinical decision-making ignores other types of evidence inherent in nursing practice (Scott-Findlay & Pollock, 2004).
To use evidence in practice, the weight of the research, also called research validity, must be determined. Evidence hierarchies have been defined to grade and assign value to the information source. For example, a classic evidential hierarchy developed by Stetler et al. (1998) prioritized evidence into six categories:
- Meta-analysis
- Individual experimental studies
- Quasi-experimental studies
- Nonexperimental studies
- Program evaluations, such as quality improvement projects
- Opinions of experts
The hierarchy identifies meta-analysis as the highest-quality evidence because it uses multiple individual research studies to reach a consensus. It is interesting to note that opinions of experts are considered the least significant in this hierarchy, yet nurses most often seek the opinion of a more experienced colleague or peer when searching for information regarding patient care (Pravikoff et al., 2005).
Qualitative research allows one to understand the way in which the intervention is experienced by the researcher and the participant and the value of the interventions to both parties (O'Neill et al., 2007). Qualitative research is not always considered in EBP because methods for synthesizing the evidence are more difficult to accomplish and interpret. The Cochrane Qualitative Research Methods Group has developed search, appraisal, and synthesis methodologies for qualitative research (Joanna Briggs Institute, n.d.) and provides a database of articles related to methodological research (Cochrane Methods, n.d.).
Bridging the Gap Between Research and Practice ⬆ ⬇
The time between research dissemination and clinical translation may be significant, and this delay may adversely affect patient outcomes. Bridging the gap between research and practice requires an understanding of the key concepts and barriers, access to research findings, access to clinical mentors for research understanding, a reinforcing culture, and a desire on the part of the clinician to implement best practices (Melnyk, 2005; Melnyk et al., 2005). In the Iowa model of EBP, research and other evidential sources are adopted directly in the practice setting with the goal of developing a standard of care and team decision-making (Schaffer et al., 2013; Titler, 2007). In addition, the groundwork required to create a conceptual framework supportive of an EBP includes workplace culture change and support of the change through leadership (Stetler et al., 1998). Beliefs and attitudes, involvement in research activities, information seeking, professional characteristics, education, and other socioeconomic factors are potential determinants of research utilization (Estabrooks et al., 2003); however, meta-analysis points out that too much original research and not enough repetition of previous studies fails to advance the knowledge base.
Developing countries are often constrained economically from accessing research sources. Organizations, such as the Cochrane Collaboration, provide free reviews to fill this void. Even so, knowledge dissemination strategies and education are required to take advantage of these resources.
In the past decade, two new research approaches have emerged that hold promise for bridging the gap between research and practice: action research and implementation science. Unlike traditional research, where the goal is to contribute to the body of nursing knowledge, these two approaches focus on studying the process of evidence as it is implemented in the clinical setting. Think of a quality improvement project with data collection and the goal of knowledge dissemination after implementation.
The National Cancer Institute sponsors an online self-paced training program titled Training Institute for Dissemination and Implementation Research in Cancer. Access it at https://cancercontrol.cancer.gov/IS/training-education/tidirc/openaccess.html. The University of Washington also sponsors the Implementation Science Resource Hub. Access it at https://impsciuw.org.
Barriers to and Facilitators of Evidence-Based Practice ⬆ ⬇
Tacia et al. (2015) concluded that there are six challenges or barriers to the application of EBP: “institutional and/or cultural barriers, lack of knowledge, lack of motivation, time management, physician and patient factors, and limited access to up to date, user-friendly technology and computer systems” (p. 93). Nurses may also see the job of interpreting research as too complex or the organizational culture as a barrier to implementation of EBP (Kieft et al., 2014; McCaughan et al., 2002). Many believe that inpatient direct care nurses lack basic knowledge of EBP and must have access to and assistance with technical resources.
Yet Melnyk et al. (2009) noted that a number of factors also facilitate the use of EBP. These driving forces include knowledge and skills in EBP; having a conviction that there is a value to using evidence in practices; and practicing in a supportive culture with tools available to sustain evidence-based care, including access to computers and databases, evidence-based content at the point of care, and the presence of EBP mentors. Tacia et al. (2015) noted that interprofessional collaboration, mentorship, and administrative support were necessary for the adoption of EBP. It is imperative that we remove the barriers and support nurses using EBP as well as continue to mentor and work with those who are just beginning to initiate EBP into their practice.
In a discussion of best practices related to the implementation of the American Association of Critical-Care Nurses' practice alert on hemodynamic monitoring, Von Rueden (2020) identified strategies for practice improvement, including a formal practice gap analysis, education using simulation, nursing rounds, and journal clubs. Interestingly, this discussion centered on a practice alert initiated in 2004 and revised in 2009 and 2016. We must find ways to be more efficient in getting research results into practice.
The Role of Informatics ⬆ ⬇
Computers are used in all areas of research: (1) literature search databases, such as CINAHL; (2) online literature reference lists, such as RefWorks; (3) data capture, collection, and coding; (4) data analysis; (5) data modeling; (6) meta-analysis; (7) qualitative analysis; and (8) dissemination of results (Saba & McCormick, 2006). The context for nursing informatics has expanded to support dramatic changes in the way science is accomplished. Information need and the collaborative component of interdisciplinary research rely heavily on technology and informatics. Technologies, such as social networking, have also improved collaboration. The use of technology and informatics in facilitating interdisciplinary and translational research is a key architectural component of the NIH's (n.d.) reengineering of the clinical research enterprise as part of its road map initiative for medical research. As technology continues to advance, so do the informatics tools available to researchers. The Institute of Translational Health Sciences (n.d.) at the University of Washington is one among many university-based research groups (at this writing in May 2020, 4,175 institutions in 147 countries) that link to REDCap (n.d.), described here:
REDCap (Research Electronic Data Capture) is a secure, web-based software platform designed to support data capture for research studies, providing 1) an intuitive interface for validated data capture; 2) audit trails for tracking data manipulation and export procedures; 3) automated export procedures for seamless data downloads to common statistical packages; and 4) procedures for data integration and interoperability with external sources. (para. 2)
This database is fully compliant with HIPAA (Health Insurance Portability and Accountability Act) and institutional review boards and supports the seamless design of online questionnaires.
Another initiative designed to support translational research and the fast-tracking of research findings into clinical practice is the National Center for Data to Health (CD2H, n.d.), part of NCATS. “By increasing collaboration, the CD2H fosters a robust translational science informatics ecosystem that collectively develops solutions to solve clinical problems faster, more efficiently, and more effectively” (CD2H, n.d., para. 2). This center has also recently added the National COVID Cohort Collaborative (N3C) in response to the pandemic.
Clinical research informatics at the Oregon Clinical and Translational Research Institute (n.d.) accelerates translational research “by providing a full suite of informatics tools for bench-to-bedside, clinical and health care systems research. Program activities span the translational spectrum, from Translational Bioinformatics to Clinical Research Informatics” (para. 1). Many other universities are utilizing informatics tools, software, and databases to engage in translational research. Translational informatics refers to the application of research informatics to translational research in order to close the gap from research to the bedside to improve the health of patients and the community.
An informatics infrastructure is critical to EBP. As early as 2008, Bakken et al. discussed expanding the context of informatics to genomic health care, shifting research paradigms, and social web technologies. Ensuring the global collaborative nature of nursing research for 2010 through 2018 required an expansion of the nursing research agenda to user information needs, data management, information support for nurses and patients, practice-based knowledge generation, and design evaluation methodologies. Giuse et al. (2005) described the evolving role of the clinical informationist (i.e., informaticist) as being a partner on the healthcare team who provides timely clinical evidence for the clinical workflow. Although not specific to nursing informatics, NCATS (2023) provides awards under its Clinical and Translational Science Awards (CTSA) program to accelerate the transfer of research to the clinical setting. The Quality and Safety Education for Nurses (QSEN, 2022) cites key competencies (i.e., knowledge, skills, and attitudes) in both EBP and informatics. With the goal of promoting the use of research findings and tool use based on these findings, the Agency for Healthcare Research and Quality (AHRQ) became an active participant in pushing evidence forward into practice. The AHRQ is a government-sponsored organization with the mission of reducing patients' risk of harm, decreasing healthcare costs, and improving patient outcomes through the promotion of research and technology applications focused on EBP. In 1999, AHRQ (2022) implemented its Translating Research into Practice (TRIP) initiative to generate knowledge about evidence-based care. In the second TRIP (TRIP-II) initiative, the focus shifted to improving health care for underserved populations and using information technology to shape translational research and health policy.
A growing number of printed and electronic resources are available to assist in creating guidelines and offering information about EBP. A selection of existing websites is shown in Table 23-1.
Table 23-1 The Role of Informatics: Online Evidence-Based ResourcesWebsite | Description |
|---|
Agency for Healthcare Research and Quality www.ahrq.gov | The Agency for Healthcare Research and Quality contains a wealth of information regarding healthcare quality. There is no charge for access to the site or its resources. |
BMJ Best Practice bestpractice.bmj.com/info/us | BMJ Best Practice takes you quickly and accurately to the latest evidence-based information, whenever and wherever you need it. Its step-by-step guidance on diagnosis, prognosis, treatment, and prevention is updated daily using robust evidence-based methodology and expert opinion. |
Orelena Hawks Puckett Institute www.puckett.org | The Orelena Hawks Puckett Institute is a not-for-profit organization engaging in activities that enhance and promote healthy child, parent, and family functioning. Its goal is to foster adoption of EBPs that build on the capacities and strengths of children, parents, families, communities, and public and private organizations. |
The Centre for Evidence-Based Medicine www.cebm.net | The Centre for Evidence-Based Medicine, located in Oxford in the United Kingdom, is devoted to developing and promoting evidence-based resources for healthcare professionals. In addition to free articles, the site also provides free teaching resources and presentations. |
CINAHL Complete www.ebsco.com/products/research-databases/cinahl-complete | CINAHL information system offers a multitude of online services, which include website link sources, CINAHL's online nursing and allied health database, document delivery, and search services. |
Cochrane www.cochrane.org | Cochrane's mission is to promote evidence-informed health decision-making by producing high-quality, relevant, accessible systematic reviews and other synthesized research evidence. Its work is internationally recognized as the benchmark for high-quality information about the effectiveness of health care. |
PubMed https://pubmed.ncbi.nlm.nih.gov | PubMed comprises more than 30 million citations for biomedical literature from MEDLINE, life science journals, and online books. Citations may include links to full-text content from PubMed Central and publishers' websites. |
Joanna Briggs Institute https://jbi.global | The Joanna Briggs Institute was established in 1996 as a resource for best care practices. Joanna Briggs was the first matron of the Adelaide Hospital in Australia and is recognized for her financial and organizational support. The Joanna Briggs Institute is a leader in developing EBPs. |
The Johns Hopkins University Evidence-Based Practice Center www.jhsph.edu/research/centers-and-institutes/johns-hopkins-evidence-based-practice-center | The Johns Hopkins University Evidence-Based Practice Center was established in 1997 and is one of 14 such centers producing comprehensive, systematic reviews for the AHRQ. |
PubMed Central www.ncbi.nlm.nih.gov/pmc | PubMed Central is a free digital archive of science-related articles managed by the National Center for Biotechnology Information. BioMed Central (an open source online archive) may be accessed here. |
Trip Medical Database www.tripdatabase.com | The Trip medical database is a clinical search tool that allows clinicians to identify the best evidence for clinical practice. |
Worldviews on Evidence-Based Nursing https://sigmapubs.onlinelibrary.wiley.com/journal/17416787 | This magazine, sponsored by Sigma Theta Tau International, is dedicated exclusively to evidence-based nursing articles. The magazine is also offered online by subscription. |
Developing Evidence-Based Practice Guidelines ⬆ ⬇
Several models have been developed to guide organizations into translating research into practice. Brief descriptions of these models are provided in Table 23-2. As an example, Titler (2007) identified the steps in the Iowa model for translating research into practice as (1) identifying the problem, issue, or topic in nursing practice; (2) research and critique of related evidence; (3) adaptation of the evidence to practice; (4) implementation of the EBP; and (5) evaluation of patient outcomes and care practices. Careful analysis and discussion of the research or other forms of evidence in this scenario may reveal that, given the context, implementation may not be practical. Following implementation, results must be monitored to determine whether the application works for the context. Thoughtful discussion of the findings will help the clinical team determine whether further research or change is warranted. As a practical application, evidence-based standards for care are developed by hospitals to meet the American Nurses Association/American Nurses Credentialing Center standards for achieving Magnet hospital recognition.
Table 23-2 Comparison of Model Approaches to Evidence-Based PracticeStetler Model (2001) | ACE Star Model (Stevens, 2002) | Iowa Model of Evidence-Based Practice to Promote Quality Care (Titler et al., 2001) |
- Preparation
- Validation
- Cooperative evaluation
- Decision-making
- Translational application
- Evaluation
| - Discovery
- Evidence summary
- Translation
- Integration
- Evaluation
| - Select the trigger as impetus for practice (knowledge- or practice-focused) change.
- Determine whether the topic is worth pursuing for the organization, and if not, pursue a new trigger.
- Determine whether there is a significant research base. If so, change; otherwise, conduct or seek more research.
- If change is appropriate for practice, implement change.
- Monitor results.
- Disseminate results.
|
Information technology is important in synthesizing the research regardless of the model. Bakken (2001) recommended (1) standardized nomenclature required for the electronic health record (standardized terminologies and structures); (2) digital sources of evidence; (3) standards that facilitate healthcare data exchange among heterogeneous systems; (4) informatics processes that support the acquisition and application of evidence to a specific clinical situation; and (5) informatics competencies (p. 1999). Bakken's recommendations encouraged the development of an infrastructure that creates a database of experiential clinical evidence.
Meta-Analysis and Generation of Knowledge ⬆ ⬇
Systematic reviews combine results from multiple primary investigations to obtain consensus on a specific area of research. Studies are discarded from the review if they are not considered sound, thereby creating a reliable end result. The strength of the systematic review is its ability to corroborate findings and reach consensus. Systematic reviews show the need for more research by revealing the areas where quantitative results may be lacking or minimal. Bias may occur if the selected studies are inadequate, all sources of evidence are not investigated, or the publications selected are not adequately diverse (Lipp, 2005). The BMJ Clinical Evidence Blog (https://blogs.bmj.com) has stressed the importance of getting evidence into health service decision-making and being cautious of evidence spin that adds bias to the reporting of the evidence.
Meta-analysis, a form of systematic review, uses statistical methods to combine the results of several studies (Cook et al., 1997). Quantitative studies are typically used. According to Glass (1976), “[M]eta-analysis is the statistical analysis of a large collection of analysis results from individual studies for the purpose of integrating the findings” (p. 3).
A typical documentation search strategy for meta-analysis begins with the identification of studies through a search of bibliographic databases, identification of meta-analysis articles that match the search criteria, elimination of those articles that do not match the search criteria, review of the reference lists in the meta-analysis for other articles that may relate to the topic, and a review of each article for quality and content. Additional sources should include unpublished works, such as conferences and dissertation abstracts, with the goal of obtaining as many relevant articles as possible. Gregson et al. (2002) identified the steps of a meta-analysis as (1) defining the problem, followed by protocol generation; (2) establishing study eligibility criteria, followed by a literature search; (3) identifying the heterogeneity of results of studies; (4) standardizing the data and statistically combining the results; and (5) conducting sensitivity testing to determine whether the combined results are the same. The often-cited criticism of meta-analysis is that the emphasis is on quantitative studies, not qualitative studies. In addition, the analysis is only as good as the studies used (Gregson et al., 2002). Collection and dissemination of these meta-analyses and systematic reviews are available in paper and on the internet, although many such databases require a subscription.
The term open access refers to a worldwide movement to make a library of knowledge available to anyone with internet access. The Open Access Initiative came about in response to the tremendous cost of research library access. Libraries pay large fees for journal subscriptions, and the richness of library references is limited to what the budget allows. The cost of keeping current with research has caused library subscriptions to decline (Yiotis, 2005). Open access adds to the controversy, with some journals charging authors for publication of their work, which in itself may provide a financial barrier to publication in this form.
According to Suber's (2004) open access overview, open access refers to digital literature that is available to anyone with internet access free of charge. There are two vehicles for open access: archives and journals. Open access journals are generally peer reviewed and freely available. The publishers of open access journals do not charge the reader but rather obtain funds for publishing elsewhere. Open access journals may charge the author or depend on other forms of funding, such as donations, grants, and advertising, to publish. SPARC (Scholarly Publishing and Academic Resources Coalition, n.d.) pointed out that most research is funded by government agencies and conducted in academic research centers; however, “[o]nce published, those that contributed to the research (from taxpayers to the institutions that supported the research itself) have to pay again to access the findings. Though research is produced as a public good, it isn't available to the public who paid for it” (para. 3). Open access promotes research collaboration and fast-tracking of results into practice.
Expanding Research Boundaries and Effects on Clinical Translation ⬆ ⬇
Thus far in this chapter, we have been focused on more traditional research methods and concepts. However, the fields of biomedical informatics and computational biology are making important contributions to our understanding of the basis of disease and allowing us to examine huge data sets searching for important insights. When the human genome project was instituted, there was a great deal of excitement in the scientific community because of the potential for understanding human biological processes and diseases. As with all research, there is still a considerable time lag before clinical change.
The future of health care is based on genomics. Bioinformatics and computational biology have provided the tools to make it possible to analyze and interpret complex biological processes. Through these developments, several projects have advanced understanding of the human genome, haplotypes, and the genomic changes related to disease. The complete sequencing of the human genome has led to the systems biology referred to as omics and has elevated scientists' ability from studying one gene or protein to studying fundamental biological processes (Box 23-1).
| Box 23-1 OMICS |
|---|
| Ome and omics are suffixes derived from the term genome. National Public Radio (2010) credits botanist Hans Winkler with merging “the Greek words ‘genesis' and ‘soma' to describe a body of genes” (para. 1) in 1920. The term genome was born from this combination, and genomics arose as the study of the genome.
Scientists describe a large-scale system or complex using the ome suffix to form words such as
|
- proteome, a collection of proteins in a cell or tissue;
- metabolome, a collection of metabolites; or
- transcriptome, a collection of RNA that has been transcribed from genes.
High-throughput analysis is critical when dealing with a large subset of data at the omic level: DNA sequences, gene expression levels, or proteins. |
The goal of the International HapMap Project (National Human Genome Research Institute [NHGRI], 2012) was to
develop a haplotype map of the human genome, the HapMap, which will describe the common patterns of human DNA sequence variation. The HapMap is expected to be a key resource for researchers to use to find genes affecting health, disease, and responses to drugs and environmental factors. The information produced by the Project will be made freely available. (para. 1)
This international partnership of scientists has taken blood samples from clusters of related people, such as parents and children, from different international regions. Using these samples, the researchers have been able to catalog some of the common variations in DNA and investigate inherited alleles. As the name implies, a haplotype map identifies a set of closely linked alleles on a chromosome, which tend to be inherited together (Figure 23-3).
Figure 23-3 International HapMap Project
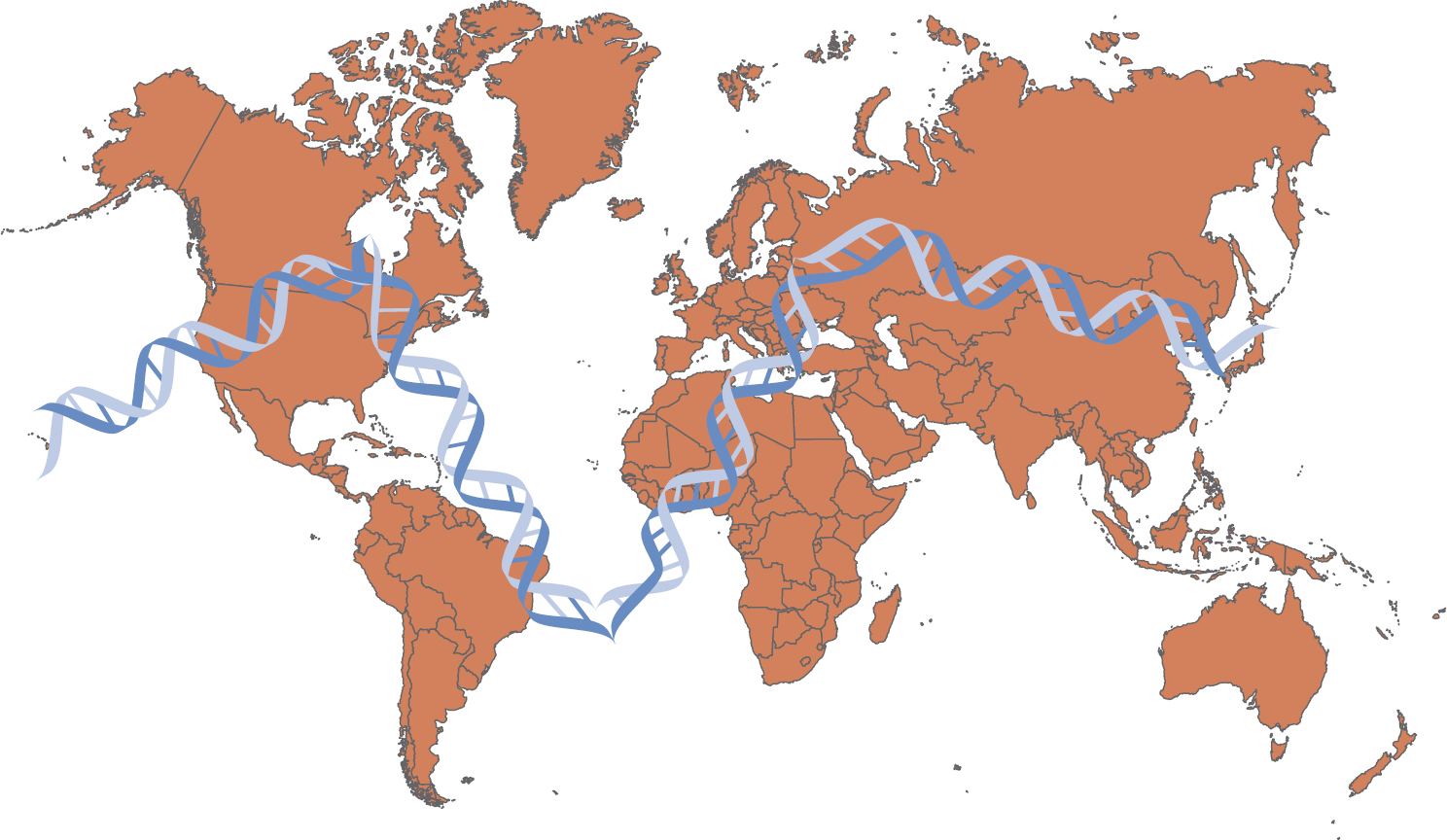
A world map features the double helix structure of D N A superimposed over North America, specifically U S A and Canada; South America, specifically Brazil; West Africa; Central Europe; and Middle Asia.
Modified from: National Human Genome Research Institute. (2012). International HapMap Project. www.genome.gov/10001688/international-hapmap-project.
The International HapMap Project states that
[m]ost common diseases, such as diabetes, cancer, stroke, heart disease, depression, and asthma, are affected by many genes and environmental factors. Although any two unrelated people are the same at about 99.9% of their DNA sequences, the remaining 0.1% is important because it contains the genetic variants that influence how people differ in their risk of disease or their response to drugs. Discovering the DNA sequence variants that contribute to common disease risk offers one of the best opportunities for understanding the complex causes of disease in humans. (para. 3)
In 2006, the Cancer Genome Atlas Program (National Cancer Institute, n.d.) began. This pilot project cost $100 million to map the genomic changes in brain, lung, and ovarian cancers to assess the feasibility of a full-scale effort to systematically explore the entire spectrum of genomic changes involved in every major type of human cancer. The goal of this project was to develop a resource that will be used to develop new strategies for preventing, diagnosing, and treating the disease.
A major contribution has been the Human Genome Project (HGP, 2019), which began in 1990 and was completed in 2003. The U.S. Department of Energy and the NIH coordinated this program, which had the following goals:
- Identify all of the approximately 20,000-25,000 genes in human DNA
- Determine the sequences of the 3 billion chemical base pairs that make up human DNA
- Store this information in databases
- Improve tools for data analysis
- Transfer related technologies to the private sector
- Address the ethical, legal, and social issues (ELSI) that may arise from the project (para. 2)
According to NHGRI (2015), “One of the most important aspects of bioinformatics is identifying genes within a long DNA sequence” (para. 1). It was clear that the speed of DNA sequencing would have to be realized sooner to decrease costs. The process was refined so that the sequencing was improved. It took 4 years to sequence the first billion bases but just 4 months to sequence the second billion bases.
During the month of January 2003, 1.5 billion bases were sequenced. As the speed of DNA sequencing increased, the cost decreased from $10 per base in 1990 to $0.10 per base at the conclusion of the project in April 2003 (NHGRI, 2015).
One of the most important aspects of bioinformatics is identifying genes within a long DNA sequence. Until the development of bioinformatics, the only way to locate genes along the chromosome was to study their function in the organism (in vivo) or to isolate the DNA and study it in a test tube (in vitro). Bioinformatics allows scientists to make educated guesses about where genes are located simply by analyzing sequence data using a computer (in silico) (NHGRI, 2015).
The other major concern brought out through the HGP was the realization that ethical, social, and legal implications (ESLI) arise from studying human genomes. The participants in the HGP set aside a percentage of their annual budgets to research ESLI (HGP, 2008). Box 23-2 and Figure 23-4 identify some of the questions raised regarding ESLI. In 1990, the NIH created a specific research program targeting ESLI and addressing human diversity, including communities and healthcare needs that may not have not been fairly considered. In 2022, the 5th Annual ESLI Congress was titled “Innovating for a Just and Equitable Future” (NHGRI, 2023).
Figure 23-4 Ethical, Social, and Legal Implications (ESLI)
A cyclic diagram elaborates on E S L I, encompassing ethical, social, and legal implications.
| Box 23-2 ESLI Questions Raised by the Human Genome Project |
|---|
- Who should have access to personal genetic information, and how will it be used?
- Who owns and controls genetic information?
- How does personal genetic information affect an individual and society's perceptions of that individual?
- How does genomic information affect members of minority communities?
- Do healthcare personnel properly counsel parents about the risks and limitations of genetic technology?
- How reliable and useful is fetal genetic testing?
- What are the larger societal issues raised by new reproductive technologies?
- How will genetic tests be evaluated and regulated for accuracy, reliability, and utility? (Currently, there is little regulation at the federal level.)
- How do we prepare healthcare professionals for the new genetics?
- How do we prepare the public to make informed choices?
- How do we as a society balance current scientific limitations and social risk with long-term benefits?
- Should testing be performed when no treatment is available?
- Should parents have the right to have their minor children tested for adult-onset diseases?
- Are genetic tests reliable and interpretable by the medical community?
- Do people's genes make them behave in a particular way?
- Can people always control their behavior?
- What is considered acceptable diversity?
- Where is the line between medical treatment and enhancement?
- Are genetically modified foods and other products safe for humans and the environment?
- How will these technologies affect developing nations' dependence on the West?
- Who owns genes and other pieces of DNA?
- Will patenting DNA sequences limit their accessibility and development into useful products?
Data from: Human Genome Project. (2008). Human Genome Project information archive, 1999-2003: Ethical, legal, and social issues. www.ornl.gov/sci/techresources/Human_Genome/elsi/elsi.shtml. |
Even though the HGP has ended, researchers continue to improve DNA sequencing. Specifically, they continue to advance the bioinformatics and computational biology tools that are used in biomedical informatics. These three projects were pivotal in genomics. The HGP focused on the DNA sequence from a single individual, the HapMap Project focused on variation in the genome and on human populations, and the Cancer Genome Atlas Project is concerned with how cancer affects the genomes. As a result of these seminal projects and a unique culture of data sharing previously unknown among biological researchers, molecular data and measurement tools are now publicly available. Two examples of publicly available databases are the Gene Expression Omnibus, which is maintained by the National Center for Biotechnology Information at the National Library of Medicine, and Array-Express, which is maintained by the European Bioinformatics Institute (Butte, 2008). As new researchers with both biology and computational expertise emerge, bioinformatics and computational biology projects will contribute new insights into disease mechanisms and therapeutic interventions, including drug repurposing. Drug repurposing means identifying new uses for previously Food and Drug Administration-approved drugs (Tang et al., 2022).
We have seen numerous advances ranging from possible treatments for Parkinson's disease and unprecedented detail of the genetics of type 2 diabetes to the NIH creating an atlas of human malformation syndromes in diverse populations, NIH researchers identifying striking genomic signatures shared by five types of cancer, and NIH scientists discovering the genetic cause of a rare allergy to vibration (NIH, 2016). However, it will take many more years of researching and applying bioinformatics and computational biology before the information in the human genome is understood in detail. Because these applications have the ability to allow one to analyze and interpret complex biological processes, researchers are on the path to understanding the etiology of disease and treatment interventions at the molecular level. Jiang et al. (2022) described the use of big data and AI processing “to facilitate biomedical breakthroughs in cancer through cross-modality integration, cross-cohort aggregation and data reuse” and urged the expansion of cancer data sets “to allow better computational models to drive basic research, cancer diagnostics and the development of new therapies” (p. 636).
Consider a typical day on any clinical unit. The advanced practice nurse who wants to prescribe a drug for a patient begins by reviewing the patient's genetic test results. The advanced practice nurse knows that this information must be assessed before prescribing so that a drug that will treat the patient's illness successfully without producing harmful side effects can be selected. The patient will receive only the medication that they need and one that is designed to interfere with or enhance the specific molecular processes that are the signature for the patient's particular health challenge. The advances that bioinformatics and biomedical informatics promise will dramatically affect healthcare delivery as it is known. As explained by Rajappa et al. (2004),
[u]nderstanding molecular mechanisms leads to better classification of disease and better management. A drop of blood from a hypertensive patient gives gene expression profile by cDNA microarray analysis. It may reveal SNPs [single nucleotide polymorphisms] related to hypertension and others which predispose a patient to diabetes mellitus or myocardial infarction and the clinician can determine which drugs are beneficial and which are harmful. This scenario has a whale of difference from the current “trial and error” method of matching a patient with antihypertensives. (p. 128)
This vision from 2004 has become a reality, and the scope of treatments and interventions continues to expand. Our expectations and hopes are being met and surpassed.
Nurses can be involved in bioinformatics in many ways, including as nurse researchers, helping to map molecular processes, and as educators and advocates, helping patients and families to understand these complex biological processes. For more information about the roles of nurses in this exciting new field, visit the website of the International Society of Nurses in Genetics (www.isong.org).
The Future ⬆ ⬇
The future depends on a prepared workforce ready to meet the challenges of tomorrow. A prepared workforce will require a focus on informatics, bioinformatics, clinical research, translational research, other research methods, and EBP. In this data-driven healthcare delivery system in which nurses work, they must adopt data standards. Given the vast amounts of data, Bakken et al. (2008) identified areas of focus for nursing informatics in knowledge representation, data management, analysis, and predictive modeling in genomic health care and the need for policies and procedures to protect data acquisition, dissemination, privacy, security, and confidentiality as well as education in these areas. Informatics tools support nursing practice, education of healthcare consumers, and knowledge generation. The technology is available now to incorporate evidence into reference links embedded in electronic clinical care plans. Incorporation of personalized clinical desktops to allow each clinician to have appropriate references (similar to internet ad bot technology) provided to them may be possible. The other challenge includes developing and maintaining interprofessional collaborative environments that truly operate in a cooperative and open manner. Time, research, and technology will tell.
Summary ⬆ ⬇
These are amazing times. Technology has taken us faster and further than we ever thought possible. Healthcare jobs have become more technical and complicated. In some ways, technology has increased the margin for error. Some healthcare practitioners will continue to rely on little scraps of paper and nonsystematic methods to keep themselves and their patients safe. Unfortunately, individuals who become so tied to these things close their mind to new innovations. The evolving quality culture and increased patient safety concerns are dragging healthcare workers forward. For the benefit of patients, health care must move forward.
The discipline of bioinformatics and its use in biomedical informatics epitomize the integration of computer science, information science, computational biology, and health care. These new applications deal with the resources, devices, strategies, and methods needed to optimize the acquisition, processing, storage, retrieval, generation, and use of information in health and biomedicine. Biomedicine and its applications of bioinformatics support and manage all healthcare behaviors. They affect how clinicians deliver health care to the infirm, prevent disease, promote health, conduct research, and provide formal education for entry-level practitioners and continuing education for those who are currently practicing. The field of biomedical informatics, which is bioinformatics capabilities coupled with health care, includes informatics and computational biology algorithms and tools and clinical guidelines. This knowledge can be applied to the areas of nursing, pharmacy, laboratory, dentistry, medicine, and public health. Those living the profession of nursing know that the practice of nursing is intertwined with the management and processing of information, including the new knowledge being generated by biomedical informatics. On the biomedical side of informatics, one must be cognizant of the fact that medical data typically are extracted from personal, confidential, and legally protected medical records. The protection of human subjects must be paramount, and all ESLI must be addressed.
Biomedical informatics provides knowledge about the effects of DNA disparities among individuals. Being able to study human genomes and biological processing at the molecular level will revolutionize how conditions are diagnosed and care is provided. The study of genomics is helping to prevent disease. If one can better understand an organism's biological processes and genetic coding, one can better prevent or treat medical conditions. Clinical care as it is known will change by becoming genomics based. Collaboration, improved access to online libraries, research tool transparency, a common data language, organizational and informational support, and continued research are a short list of items needed to advance translational research. Repeat studies are needed to provide meaningful meta-analysis and systematic reviews. Technology advancement in the area of incorporating evidence into clinical tools must continue. Removing the barriers to knowledge-seeking behavior and providing access to evidential resources will promote knowledge and, in the end, improve patient outcomes.
| Thought-Provoking Questions |
|---|
- Twelve-hour shifts are problematic for patients' and nurses' safety, yet hospitals continue to keep the 12-hour shift schedule. In 2004, the Institute of Medicine (2004) published a report that referred to studies as early as 1988 that discussed the negative effects of rotating shifts on intervention accuracy. Workers with 12-hour shifts experienced more fatigue than workers on 8-hour shifts. In another study done in Turkey by Ilhan et al. (2006), factors relating to increased risk for injury were age of 24 years or younger, less than 4 years of nursing experience, working in surgical intensive care units, and working for more than 8 hours. As a clinician reading these studies, what would your next step be?
- The use of heparin versus saline to maintain the patency of peripheral intravenous catheters has been addressed in research for many years. The American Society of Health System Pharmacists published a position paper in January 2006 advocating its support of the use of 0.9% saline in the maintenance of peripheral catheters in nonpregnant adults. It seems surprising that this position paper references articles that advocate the use of saline over heparin dating from 1991. What do you believe are some of the barriers that would have caused this delay in implementation?
- In the era of EBP, healthcare providers must continue to think critically about their actions. What is the science behind their interventions? Healthcare workers must no longer do things one way just because they have always been done that way. Research the problem, use evidence-based resources, critically select electronic and nonelectronic references, consolidate the research findings and combine and compare the conclusions, present the findings, and propose a solution. The nurse will be the first to ask why and may be a key player in making change happen.
- After reading this chapter, you know that the study of genomics is helping clinicians to understand better the interaction between genes and the environment. This new information and knowledge will continue to help clinicians find ways to improve health and prevent disease. How do you envision patient care will change based on genomics in 10 years, 20 years, or 50 years into the future?
- Review the ESLI raised by the HGP that are presented in Box 23-2. Prepare a similar list of ESLI questions to apply to the public health databases being developed for health information exchanges. Can you appreciate how these ESLI questions are widely applicable to protecting information gathered from human subjects?
|
References ⬆
- Aarden E., Marelli L., & Blasimme A. (2021). The translational lag narrative in policy discourse in the United States and the European Union: A comparative study. Humanities & Social Sciences Communications, 8, 107. https://doi.org/10.1057/s41599-021-00777-y
- Agency for Healthcare Research and Quality. (2022). Agency for Healthcare Research and Quality: A profile. www.ahrq.gov/cpi/about/profile/index.html
- American Medical Informatics Association. (n.d.-a). Areas of practice: Clinical informatics. www.amia.org/applications-informatics/clinical-informatics
- American Medical Informatics Association. (n.d.-b). Areas of practice: Clinical research informatics. www.amia.org/applications-informatics/clinical-research-informatics
- American Medical Informatics Association. (n.d.-c). Areas of practice: Translational bioinformatics. www.amia.org/applications-informatics/translational-bioinformatics
- American Society of Health System Pharmacists. (2006). ASHSP therapeutic position statement on the institutional use of 0.9% sodium chloride injection to maintain patency of peripheral indwelling intermittent infusion devices.American Journal of Health System Pharmacy, 63(13), 1273-1275. https://doi.org/10.2146/ajhp060094
- Bakken S. (2001). An informatics infrastructure is essential for evidence-based practice. Journal of the American Medical Informatics Association, 8(3), 199-201. https://doi.org/10.1136/jamia.2001.0080199
- Bakken S., Stone P. W., & Larson E. L. (2008). A nursing informatics research agenda for 2008-18: Contextual influences and key components. Nursing Outlook, 56(5), 206-214. https://doi.org/10.1016/j.outlook.2008.06.007
- Baumann S. (2010). The limitations of evidence-based practice. Nursing Science Quarterly, 23(3), 226-230. https://doi.org/10.1177/0894318410371833
- Brown S. (2013). Evidence-based nursing: The research-practice connection. Jones & Bartlett Learning.
- Butte A. (2008). Translational bioinformatics: Coming of age. Journal of the American Medical Informatics Association, 15(6), 709-714. https://doi.org/10.1197/jamia.M2824
- Cochrane A. L. (1972). Effectiveness and efficiency: Random reflections on health services. Nuffield Provincial Hospitals Trust.
- Cochrane Methods. (n.d.). Methodology register. https://methods.cochrane.org/qi/methodology-register
- Cook D. J., Mulrow C. D., & Haynes R. B. (1997). Systematic reviews: Synthesis of best evidence for clinical decisions. Annals of Internal Medicine, 126(5), 376-380. https://doi.org/10.7326/0003-4819-126-5-199703010-00006
- Dickersin K., & Manheimer E. (1998). The Cochrane Collaboration: Evaluation of health care services using systematic reviews of the results and randomized controlled trials. Clinical Obstetrics and Gynecology, 41(2), 315-331. https://doi.org/10.1097/00003081-199806000-00012
- Estabrooks C. A., Floyd J. A., Scott-Findlay S., O'Leary K. A., & Gushta M. (2003). Individual determinants of research utilization: A systematic review. Journal of Advanced Nursing, 43(5), 73-81. https://doi.org/10.1046/j.1365-2648.2003.02748.x
- Fawcett J., Watson J., Walker P. H., & Fitzpatrick J. J. (2001). On nursing theories and evidence. Journal of Nursing Scholarship, 33(2), 115-119. https://doi.org/10.1111/j.1547-5069.2001.00115.x
- French P. (2002). What is the evidence on evidence-based nursing? An epistemological concern. Journal of Advanced Nursing, 37(3), 250-257. https://doi.org/10.1046/j.1365-2648.2002.02065.x
- Giuse N. B., Koonce T. Y., Jerome R. N., Gahall M., Sathe N. A., & Williams A. (2005). Evolution of a mature clinical informationist model. Journal of the American Medical Informatics Association, 12(3), 249-255. https://doi.org/10.1197/jamia.M1726
- Glass G. V. (1976). Primary, secondary, and meta-analysis of research. Educational Research, 5(10), 3-8. https://doi.org/10.3102/0013189X005010003
- Goode C. J., & Piedalue F. (1999). Evidence-based clinical practice. Journal of Nursing Administration, 29(6), 15-21. https://doi.org/10.1097/00005110-199906000-00005
- Gregson P. R., Meal A. G., & Avis M. (2002). Meta-analysis: The glass eye of evidence-based practice? Nursing Inquiry, 9(1), 24-30. https://doi.org/10.1046/j.1440-1800.2002.00129.x
- Human Genome Project. (2008). Human Genome Project information archive, 1990-2003: Ethical, legal, and social issues. www.ornl.gov/sci/techresources/Human_Genome/elsi/elsi.shtml
- Human Genome Program. (2019). Human Genome Project information archive, 1990-2003. http://web.ornl.gov/sci/techresources/Human_Genome
- Ilhan M. N., Durukan E., Aras E., Turkcuoglu S., & Aygun R. (2006). Long working hours increase the risk of sharp and needlestick injury in nurses: A need for new policy implication. Journal of Advanced Nursing, 56(5), 563-568. https://doi.org/10.1111/j.1365-2648.2006.04041.x
- Institute of Medicine. (2004). Keeping patients safe. National Academies Press.
- Institute of Translational Health Sciences. (n.d.). REDCap. www.iths.org/investigators/services/bmi/redcap
- Jiang P., Sinha S., Aldape K., Hannenhalli S., Sahinalp C., & Ruppin E. (2022). Big data in basic and translational cancer research. Nature Reviews Cancer, 22(11), 625-639. https://doi.org/10.1038/s41568-022-00502-0
- Joanna Briggs Institute. (n.d.). Home page. https://joannabriggs.org
- Kieft R., de Brouwer B., Francke A., & Delnoij D. (2014). How nurses and their work environment affect patient experiences of the quality of care: A qualitative study. BMC Health Services Research, 14, 249. https://doi.org/10.1186/1472-6963-14-249
- Kirchhoff K. T. (2004). State of the science of translational research: From demonstration projects to intervention testing. Worldviews on Evidence-Based Nursing, 1(S1), S6-S12. https://doi.org/10.1111/j.1524-475X.2004.04039.x
- Lipp A. (2005). The systematic review as an evidence-based tool for the operating room. AORN Journal, 81(6), 1279-1287. https://doi.org/10.1016/s0001-2092(06)60393-1
- McCaughan D., Thompson C., Cullum N., Sheldon T. A., & Thompson D. R. (2002). Acute care nurses' perceptions of barriers to using research information in clinical decision-making. Journal of Advanced Nursing, 39(1), 46-60. https://doi.org/10.1046/j.1365-2648.2002.02241.x
- Melnyk B. M. (2005). Advancing evidence-based practice in clinical and academic settings. Worldviews on Evidence-Based Nursing, 2(3), 161-165. https://doi.org/10.1111/j.1741-6787.2005.00027.x
- Melnyk B. M., Fineout-Overholt E., Stetler C., & Allen J. (2005). Outcomes and implementation strategies from the first U.S. evidence-based practice leadership summit. Worldviews on Evidence-Based Nursing, 2(3), 113-121. https://doi.org/10.1111/j.1741-6787.2005.00022.x
- Melnyk B. M., Fineout-Overholt E., Stillwell S., & Williamson K. (2009). Igniting a spirit of inquiry: An essential foundation for evidence-based practice. American Journal of Nursing, 109(11), 49-52. https://doi.org/10.1097/01.NAJ.0000363354.53883.58
- Melnyk B. M., Fineout-Overholt E., Stone P., & Ackerman M. (2000). Evidence-based practice: The past, the present, and recommendations for the millennium. Pediatric Nursing, 26(1), 77-80.
- National Cancer Institute. (n.d.). The Cancer Genome Atlas program. www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga
- National Center for Advancing Translational Sciences. (2019). Transforming translational science.https://ncats.nih.gov/files/NCATS_Factsheet_508.pdf
- National Center for Advancing Translational Sciences. (2021). Translational science spectrum. https://ncats.nih.gov/translation/spectrum
- National Center for Advancing Translational Sciences. (2023). Clinical and Translational Science Awards (CTSA) program. www.ncats.nih.gov/ctsa
- National Center for Data to Health. (n.d.). CD2H: Harmonizing the informatics community. https://cd2h.org
- National Human Genome Research Institute. (2012). International HapMap Project. www.genome.gov/10001688/international-hapmap-project
- National Human Genome Research Institute. (2015). Bioinformatics: Finding genes. www.genome.gov/25020001/online-education-kit-bioinformatics-finding-genes
- National Human Genome Research Institute. (2023). Ethical, legal, and social implications research program. www.genome.gov/Funded-Programs-Projects/ELSI-Research-Program-ethical-legal-social-implications#areas
- National Institutes of Health. (n.d.). The NIH roadmap: Re-engineering the clinical research enterprise. http://or.org/pdf/NIH_Roadmap-ClinicalResearch.pdf
- National Institutes of Health. (2016, February 3). NIH scientists discover genetic cause of rare allergy to vibration.www.nih.gov/news-events/news-releases/nih-scientists-discover-genetic-cause-rare-allergy-vibration
- National Public Radio. (2010, July 9). Where the word “genome” came from. www.npr.org/templates/story/story.php?storyId=128410577
- O'Neill T., Jinks C., & Ong B. N. (2007). Decision-making regarding total knee replacement surgery: A qualitative meta-synthesis. BMC Health Services Research, 7(52). https://doi.org/10.1186/1472-6963-7-52
- Oregon Clinical and Translational Research Institute. (n.d.). Powering innovation with OCTRI informatics. www.ohsu.edu/octri/powering-innovation-state-art-informatics
- Polit D. F., & Beck T. C. (2008). Nursing research: Generating and assessing evidence for nursing practice (8th ed.). Lippincott Williams & Wilkins.
- Pravikoff D. S., Tanner A. B., & Pierce S. T. (2005). Readiness of U.S. nurses for evidence-based practice. American Journal of Nursing, 105(9), 40-51. https://doi.org/10.1097/00000446-200509000-00025
- Quality and Safety Education for Nurses. (2022). Evidence-based practice (EBP). www.qsen.org/post/evidence-based-practice
- Rajappa M., Sharma A., & Saxena A. (2004). Bioinformatics and its implications in clinical medicine: A review. International Medical Journal, 11(2), 125-129.
- REDCap. (n.d.). Citations. www.project-redcap.org/resources/citations
- Rycroft-Malone J., Seers K., Titchen A., Harvey G., Kitson A., & McCormack B. (2004). What counts as evidence in evidence-based practice? Journal of Advanced Nursing, 47(1), 81-90. https://doi.org/10.1111/j.1365-2648.2004.03068.x
- Saba V. K., & McCormick K. A. (2006). Essentials of nursing informatics (4th ed.). McGraw-Hill.
- Sackett D. I., Rosenberg W. M., Gray J. A., Haynes R. B., & Richardson W. S. (1996). Evidence based medicine: What it is and what it isn't. British Medical Journal, 312, 71-72. https://doi.org/10.1136/bmj.312.7023.71
- Schaffer M., Sandau K., & Diedrick L. (2013). Evidence-based practice models for organizational change: Overview and practical applications. Journal of Advanced Nursing, 69(5), 1197-1209. https://doi.org/10.1111/j.1365-2648.2012.06122.x
- Scholarly Publishing and Academic Resources Coalition. (n.d.). Open access. https://sparcopen.org/open-access
- Scott-Findlay S., & Pollock C. (2004). Evidence, research, knowledge: A call for conceptual clarity. Worldviews on Evidence-Based Nursing, 1(2), 92-97. https://doi.org/10.1111/j.1741-6787.2004.04021.x
- Squires J. E., Estabrooks C. A., Gustavsson P., & Wallin L. (2011). Individual determinants of research utilization by nurses: A systematic review update. Implementation Science, 6(1). https://doi.org/10.1186/1748-5908-6-1
- Stetler C. B. (2001). Updating the Stetler model of research utilization to facilitate evidence-based practice. Nursing Outlook, 49(6), 272-279. https://doi.org/10.1067/mno.2001.120517
- Stetler C. B., Brunell M., Giuliano K. K., Morse D., Prince L., & Newell-Stokes V. (1998). Evidence-based practice and the role of nursing leadership. Journal of Nursing Administration, 28(7/8), 45-53. https://doi.org/10.1097/00005110-199807000-00011
- Stevens K. R. (2002). ACE star model of the cycle of knowledge transformation. http://sharonbsn.tripod.com/new_page_1.htm
- Suber P. (2004, December 29). A very brief introduction to open access. http://legacy.earlham.edu/~peters/fos/brief.htm
- Tacia L., Biskupski K., Pheley A., & Lehto R. (2015). Identifying barriers to evidence-based practice adoption: A focus group study. Clinical Nursing Studies, 3(2), 90-96. https://doi.org/10.5430/cns.v3n2p90
- Tang A. A., Woldemariam S. S., Roger J. J., & Sirota M. M. (2022). Translational bioinformatics to enable precision medicine for all: Elevating equity across molecular, clinical, and digital realms. Yearbook of Medical Informatics, 31(1), 106-115. https://doi.org/10.1055/s-0042-1742513
- Titler M. G. (2004). Methods in translation science. Worldviews on Evidence-Based Nursing, 1(1), 38-48. https://doi.org/10.1111/j.1741-6787.2004.04008.x
- Titler M. G. (2007). Translating research into practice. American Journal of Nursing, 107(6), 26-33. https://doi.org/10.1097/01.NAJ.0000277823.51806.10
- Titler M. G. (2010). Translation science and context. Research and Theory for Nursing Practice, 24(1), 35-55. http://web.archive.org/web/20220709104946/https://connect.springerpub.com/content/sgrrtnp/24/1/35
- Titler M. G., Kleiber C., Steelman V., Rakel B., Budreu G., Everett L., Buckwalter K. C., Tripp-Reimer T., & Goode T. (2001). The Iowa model of evidence-based practice to promote quality care. Critical Care Nursing Clinics of North America, 13(4), 497-509. https://doi.org/10.1016/S0899-5885(18)30017-0
- Von Rueden K. T. (2020). Bridging the gap between clinical practice and the AACN practice alert on pulmonary artery/central venous pressure monitoring in adults. AACN Advanced Critical Care, 31(1), 34-40. https://doi.org/10.4037/aacnacc2020888
- Weiss M. E., Bobay K. L., Johantgen M., & Shirey M. R. (2018). Aligning evidence-based practice with translational research. Journal of Nursing Administration, 48(9), 425-431. https://doi.org/10.1097/NNA.0000000000000644
- Woolf S. H. (2008). The meaning of translational research and why it matters. Journal of the American Medical Association, 299(2), 211-213. https://doi.org/10.1001/jama.2007.26
- Yiotis K. (2005). The Open Access Initiative: A new paradigm for scholarly communications. Information Technology and Libraries, 24(4), 157-162. https://doi.org/10.6017/ital.v24i4.3378




